produce a barplot of cell barcode demultiplex statistics
Usage
plot_demultiplex(pipeline)
# S4 method for class 'FLAMES.SingleCellPipeline'
plot_demultiplex(pipeline)Value
a list of ggplot objects:
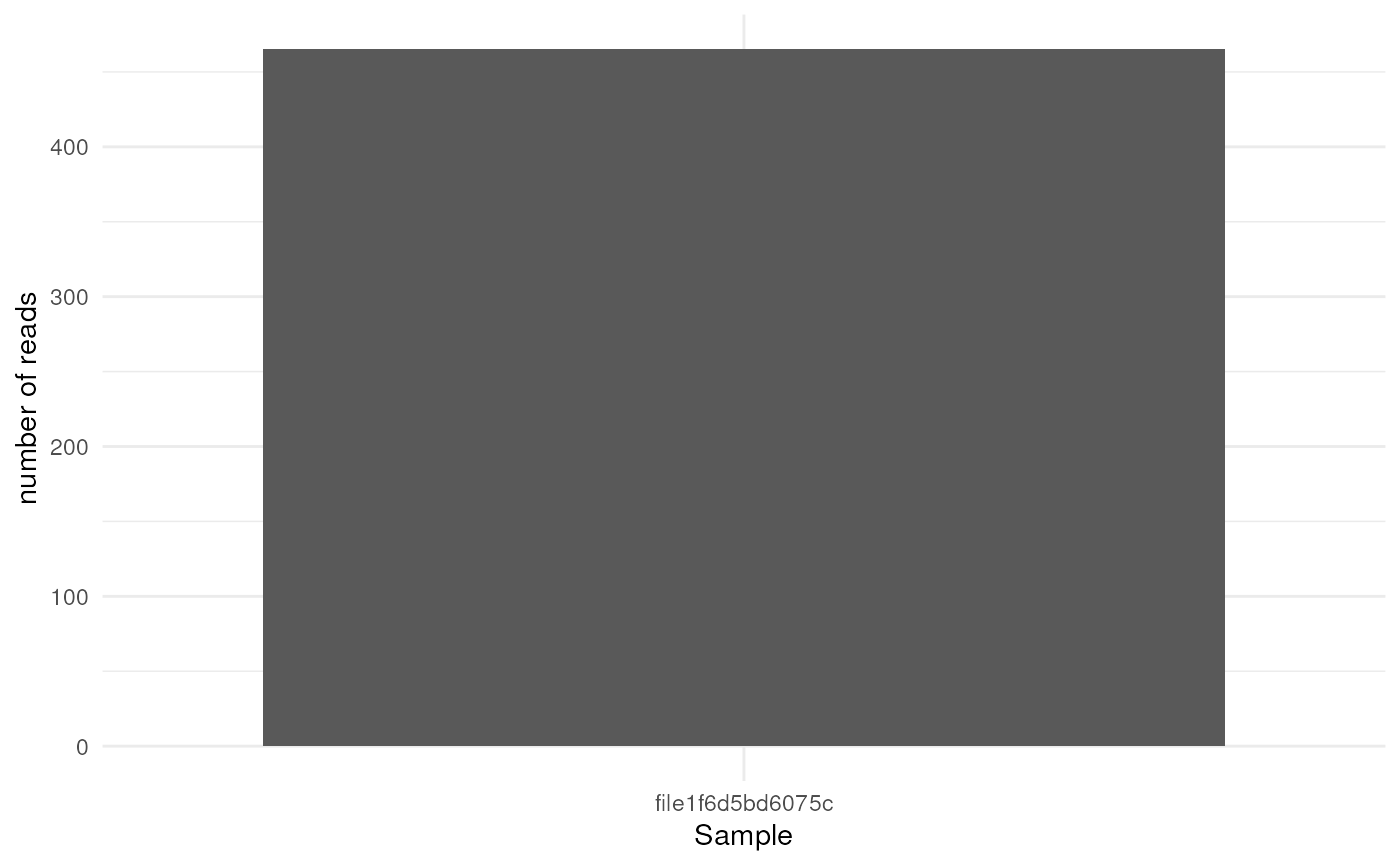
reads_count_plot: stacked barplot of: demultiplexed reads
knee_plot: knee plot of UMI counts before TSO trimming
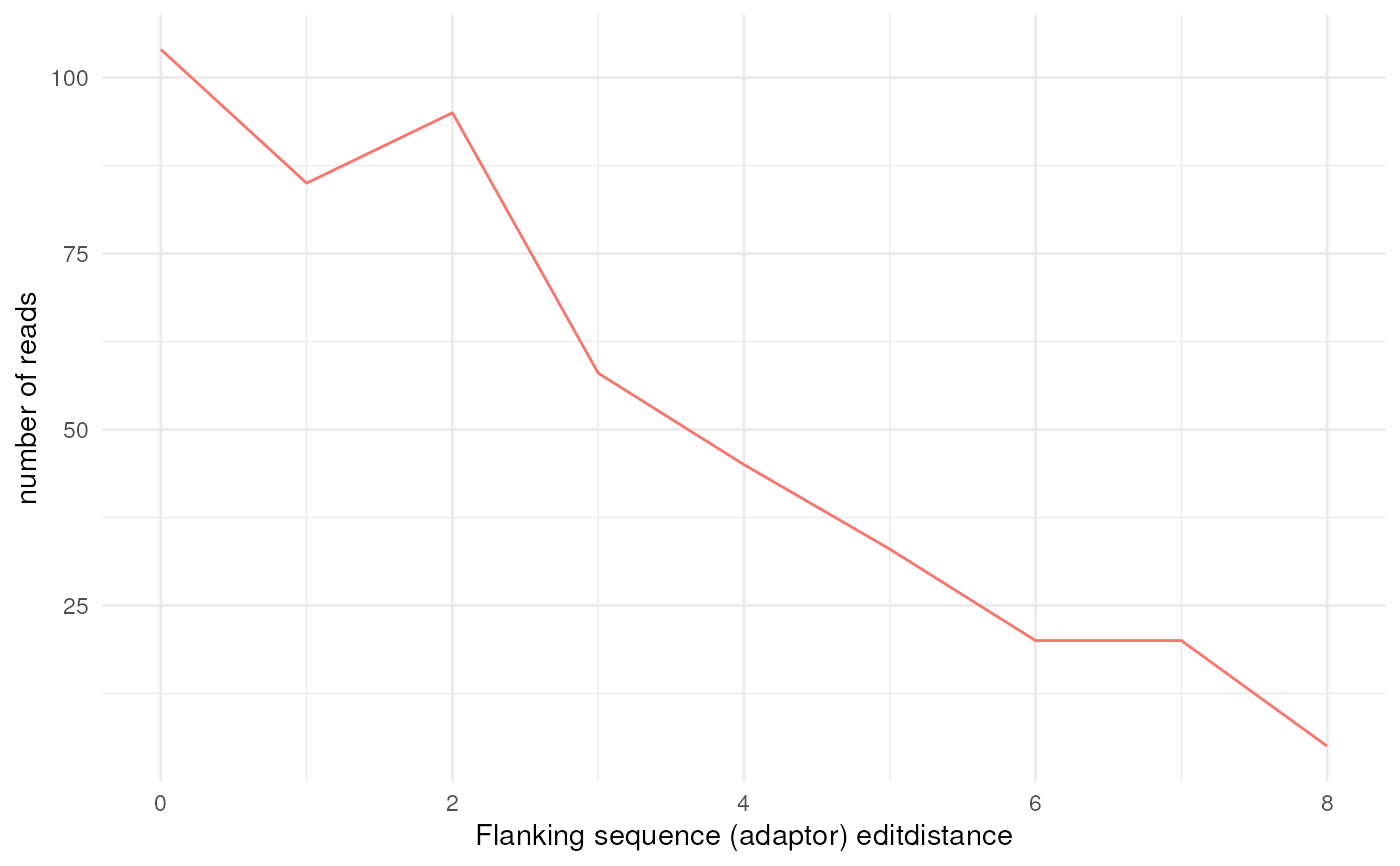
flank_editdistance_plot: flanking sequence (adaptor) edit-distance plot
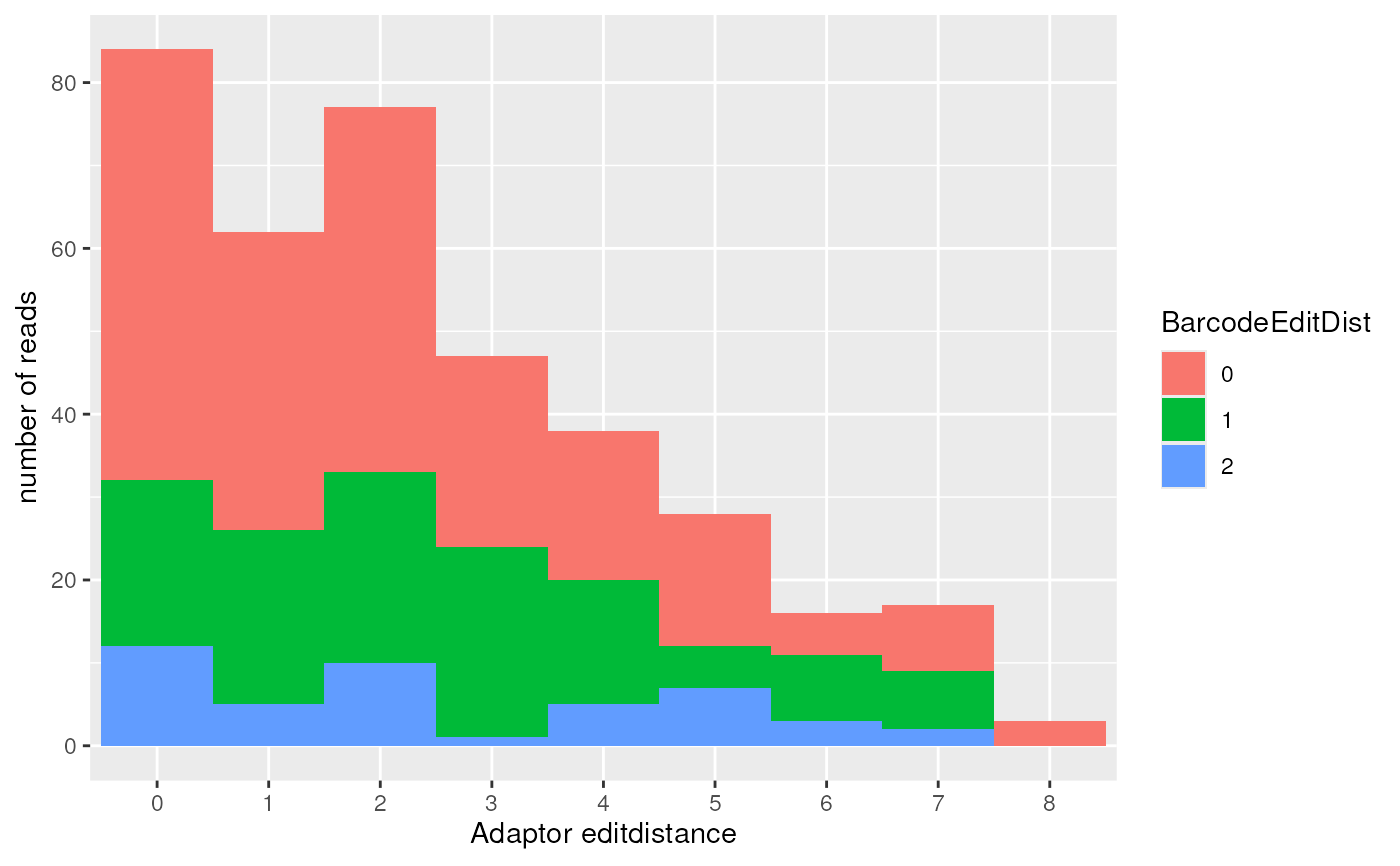
barcode_editdistance_plot: barcode edit-distance plot
cutadapt_plot: if TSO trimming is performed, number of reads kept by cutadapt
Examples
pipeline <- example_pipeline("MultiSampleSCPipeline") |>
run_step("barcode_demultiplex")
#> Writing configuration parameters to: /tmp/RtmpmJ8vO7/file80d05e2b564b/config_file_32976.json
#> Configured steps:
#> barcode_demultiplex: TRUE
#> genome_alignment: TRUE
#> gene_quantification: TRUE
#> isoform_identification: TRUE
#> read_realignment: TRUE
#> transcript_quantification: TRUE
#> samtools not found, will use Rsamtools package instead
#> ── Running step: barcode_demultiplex @ Fri Oct 31 06:51:52 2025 ────────────────
#> Using flexiplex for barcode demultiplexing.
#> FLEXIPLEX 0.96.2
#> Setting max barcode edit distance to 2
#> Setting max flanking sequence edit distance to 8
#> Setting read IDs to be replaced
#> Setting number of threads to 1
#> Search pattern:
#> primer: CTACACGACGCTCTTCCGATCT
#> BC: NNNNNNNNNNNNNNNN
#> UMI: NNNNNNNNNNNN
#> polyT: TTTTTTTTT
#> Setting known barcodes from /tmp/RtmpmJ8vO7/file80d05e2b564b/bc_allow.tsv
#> Number of known barcodes: 143
#> Processing file: /tmp/RtmpmJ8vO7/file80d05e2b564b/fastq/sample1.fq.gz
#> Searching for barcodes...
#> Processing file: /tmp/RtmpmJ8vO7/file80d05e2b564b/fastq/sample2.fq.gz
#> Searching for barcodes...
#> Processing file: /tmp/RtmpmJ8vO7/file80d05e2b564b/fastq/sample3.fq.gz
#> Searching for barcodes...
#> Number of reads processed: 393
#> Number of reads where at least one barcode was found: 368
#> Number of reads with exactly one barcode match: 364
#> Number of chimera reads: 1
#> All done!
#> Reads Barcodes
#> 10 2
#> 9 2
#> 8 5
#> 7 4
#> 6 3
#> 5 7
#> 4 14
#> 3 14
#> 2 29
#> 1 57
#> FLEXIPLEX 0.96.2
#> Setting max barcode edit distance to 2
#> Setting max flanking sequence edit distance to 8
#> Setting read IDs to be replaced
#> Setting number of threads to 1
#> Search pattern:
#> primer: CTACACGACGCTCTTCCGATCT
#> BC: NNNNNNNNNNNNNNNN
#> UMI: NNNNNNNNNNNN
#> polyT: TTTTTTTTT
#> Setting known barcodes from /tmp/RtmpmJ8vO7/file80d05e2b564b/bc_allow.tsv
#> Number of known barcodes: 143
#> Processing file: /tmp/RtmpmJ8vO7/file80d05e2b564b/fastq/sample1.fq.gz
#> Searching for barcodes...
#> Number of reads processed: 100
#> Number of reads where at least one barcode was found: 92
#> Number of reads with exactly one barcode match: 91
#> Number of chimera reads: 1
#> All done!
#> Reads Barcodes
#> 4 1
#> 3 9
#> 2 9
#> 1 44
#> FLEXIPLEX 0.96.2
#> Setting max barcode edit distance to 2
#> Setting max flanking sequence edit distance to 8
#> Setting read IDs to be replaced
#> Setting number of threads to 1
#> Search pattern:
#> primer: CTACACGACGCTCTTCCGATCT
#> BC: NNNNNNNNNNNNNNNN
#> UMI: NNNNNNNNNNNN
#> polyT: TTTTTTTTT
#> Setting known barcodes from /tmp/RtmpmJ8vO7/file80d05e2b564b/bc_allow.tsv
#> Number of known barcodes: 143
#> Processing file: /tmp/RtmpmJ8vO7/file80d05e2b564b/fastq/sample2.fq.gz
#> Searching for barcodes...
#> Number of reads processed: 100
#> Number of reads where at least one barcode was found: 95
#> Number of reads with exactly one barcode match: 94
#> Number of chimera reads: 0
#> All done!
#> Reads Barcodes
#> 4 2
#> 3 3
#> 2 16
#> 1 47
#> FLEXIPLEX 0.96.2
#> Setting max barcode edit distance to 2
#> Setting max flanking sequence edit distance to 8
#> Setting read IDs to be replaced
#> Setting number of threads to 1
#> Search pattern:
#> primer: CTACACGACGCTCTTCCGATCT
#> BC: NNNNNNNNNNNNNNNN
#> UMI: NNNNNNNNNNNN
#> polyT: TTTTTTTTT
#> Setting known barcodes from /tmp/RtmpmJ8vO7/file80d05e2b564b/bc_allow.tsv
#> Number of known barcodes: 143
#> Processing file: /tmp/RtmpmJ8vO7/file80d05e2b564b/fastq/sample3.fq.gz
#> Searching for barcodes...
#> Number of reads processed: 193
#> Number of reads where at least one barcode was found: 181
#> Number of reads with exactly one barcode match: 179
#> Number of chimera reads: 0
#> All done!
#> Reads Barcodes
#> 7 1
#> 6 1
#> 5 1
#> 4 7
#> 3 10
#> 2 27
#> 1 53
plot_demultiplex(pipeline)
#> $reads_count_plot
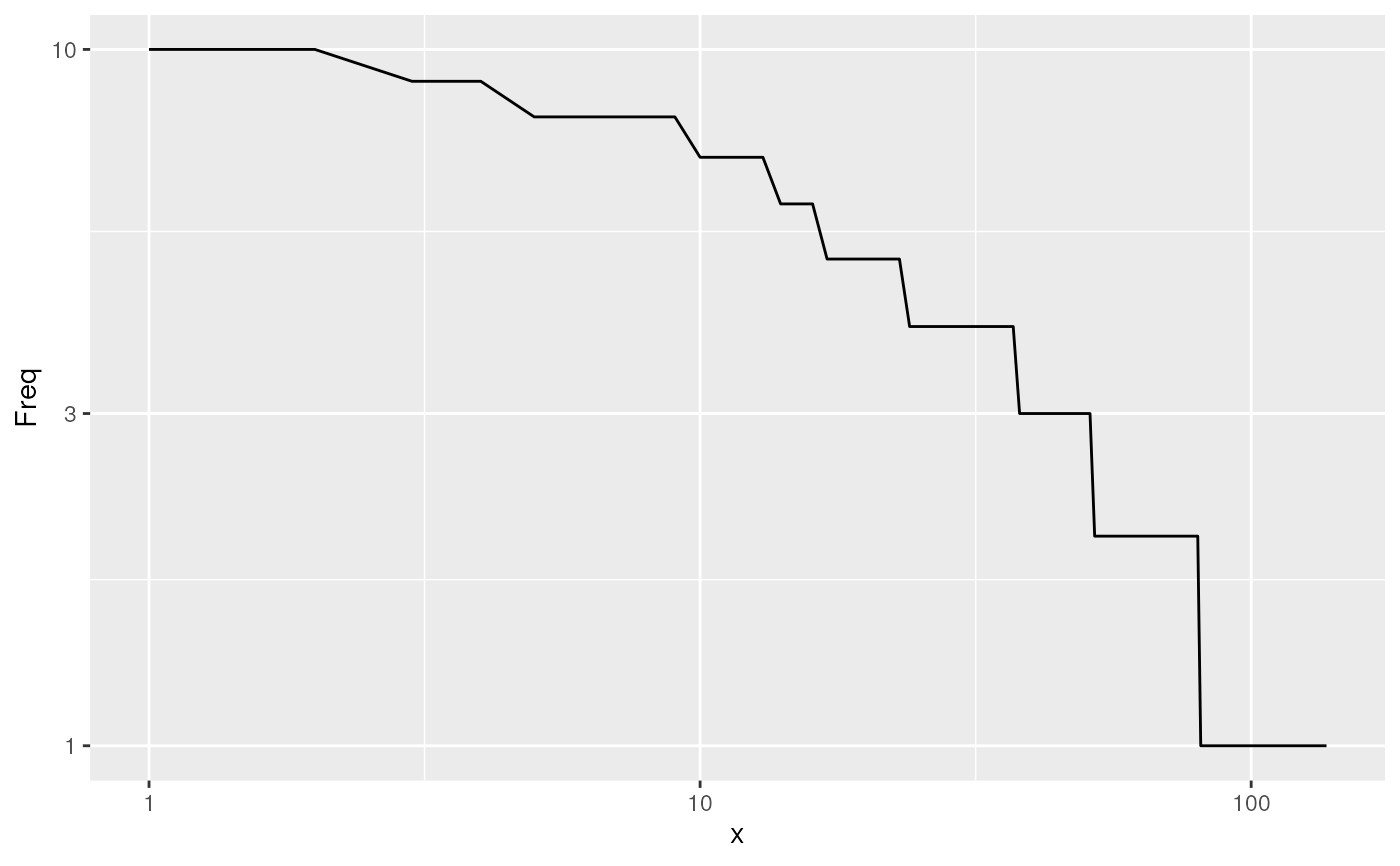 #>
#> $knee_plot
#> `geom_smooth()` using formula = 'y ~ x'
#>
#> $knee_plot
#> `geom_smooth()` using formula = 'y ~ x'
 #>
#> $flank_editdistance_plot
#>
#> $flank_editdistance_plot
 #>
#> $barcode_editdistance_plot
#>
#> $barcode_editdistance_plot
 #>
#> $cutadapt_plot
#>
#> $cutadapt_plot
 #>
#>