A streamlined workflow for long-read DNA methylation analysis with NanoMethViz and Bioconductor
Shian Su1, Lucinda Xiao1, James Lancaster1, Tamara Cameron1, Kelsey Breslin1, Peter F. Hickey1, Marnie E. Blewitt1, Quentin Gouil1, Matthew E. Ritchie1
Source:vignettes/workflow.rmd
workflow.rmd1The Walter and Eliza Hall Institute of Medical Research, Parkville, Victoria, Australia and Department of Medical Biology, The University of Melbourne, Parkville, Victoria, Australia.
Abstract
Long-read sequencing technologies have transformed the field of
epigenetics by enabling direct, single-base resolution detection of DNA
modifications, such as methylation. This produces novel opportunities
for studying the role of DNA methylation in gene regulation, imprinting,
and disease. However, the unique characteristics of long-read data,
including the modBAM format and extended read lengths, necessitate the
development of specialised software tools for effective analysis. The
NanoMethViz package provides a suite of tools for loading
in long-read methylation data, visualising data at various data
resolutions. It can convert the data for use with other Bioconductor
software such as bsseq, DSS,
dmrseq and edgeR to discover differentially
methylated regions (DMRs).
In this workflow article, we demonstrate the process of converting modBAM files into formats suitable for comprehensive downstream analysis. We leverage NanoMethViz to conduct an exploratory analysis, visually summarizing differences between samples, examining aggregate methylation profiles across gene and CpG islands, and investigating methylation patterns within specific regions at the single-read level. Additionally, we illustrate the use of dmrseq for identifying DMRs and show how to integrate these findings into gene-level visualization plots. Our analysis is applied to a triplicate dataset of haplotyped long-read methylation data from mouse neural stem cells, allowing us to visualize and compare the characteristics of the parental alleles on chromosome 7. By applying DMR analysis, we recover DMRs associated with known imprinted genes and visualise the methylation patterns of these genes summarised at single-read resolution. Through DMR analysis, we identify DMRs associated with known imprinted genes and visualize their methylation patterns at single-read resolution. This streamlined workflow is adaptable to common experimental designs and offers flexibility in the choice of upstream data sources and downstream statistical analysis tools.
Introduction
DNA methylation is a key epigenetic regulator of gene expression in mammals, involving the addition of a methyl group to cytosine or adenine nucleotides. In mammals, the predominant form of DNA methylation is 5-methylcytosine (5mC) in CpG dinucleotides. CpG methylation is involved in many important epigenetic processes, including differential gene expression, X-chromosome inactivation, genomic imprinting, and maintenance of genome stability (1). Changes in CpG methylation are implicated in a variety of different disease processes, including tumorigenesis and imprinting disorders such as Prader-Willi Syndrome and Angelman Syndrome (2).
Over the past couple of decades, multiple methods have been developed to measure DNA methylation, including bisulfite sequencing (3), methylation arrays (4), and more recently, third-generation sequencing technologies such as those offered by Oxford Nanopore Technologies (ONT) and Pacific Biosciences (PacBio). While bisulfite sequencing has long been the gold standard for DNA methylation analysis, it suffers from several limitations; these include DNA degradation from the process of bisulfite conversion, incomplete conversion of unmethylated cytosines, amplification bias from PCR, and short read lengths that pose difficulties for assembly and haplotyping (5). By contrast, ONT’s nanopore sequencing and PacBio’s single-molecule real-time (SMRT) sequencing can directly sequence DNA molecules with all their modifications intact, and use basecalling algorithms to predict DNA modifications from the raw sequencing signal. Basecalling algorithms have the potential for bias, depending on the algorithm and training data used for the model; however, improvements are continually being made to generate more accurate models, and additionally, to detect other DNA modifications in other sequence contexts (6).
In this workflow, we will demonstrate how to use the
NanoMethViz package (7) and
other Bioconductor software to visualise and analyse methylation data
generated by ONT and PacBio long-read sequencing. The sequencing data is
currently output in the modBAM file format, which contains the
basecalled reads alongside tags for modification information. The
NanoMethViz package provides a suite of tools for loading
in this methylation data, visualising regional and genome-wide
methylation patterns, and exporting data for identifying differentially
methylated regions (DMRs) using other Bioconductor software such as
bsseq (8), DSS
(9), dmrseq (10) and edgeR (11) (Figure @ref(fig:fig1)B).
Experimental Setup
In this workflow, we will analyse ultra-long-read Nanopore sequencing data from the neural stem cells of triplicate E14.5 female mice (12). The mice were F1 crosses between an Xist knockout (XΔAX) female (13) and a Castaneus strain (CAST) male. The resulting offspring have sufficiently different parental chromosomes to allow effective genome-wide haplotyping to distinguish the parent-of-origin for long-reads, allowing for the identification of genes that are imprinted or strain-specific. The Xist KO of the maternal genome also guarantees that X-inactivation takes place on the paternal X chromosome, allowing for the study of X-inactivation. The DNA was sequenced on R9.4.1 PromethION flow cells, basecalled with Dorado, aligned to the mm10 mouse reference genome, and phased with WhatsHap (14), to produce the phased modBAM (Figure @ref(fig:fig1)A). For illustrative purposes, we use a we use a subset of the data focusing on chromosome 7, which contains several known imprinted genes.
.](Figure1.png)
Overview of the workflow. (A) Experimental setup for
the data used in this workflow, from Gocuk and Lancaster et al., 2024.
(B) Flowchart of the workflow for long-read methylation data
visualisation and analysis using NanoMethViz and other
Bioconductor packages including limma, dmrseq,
bsseq, and edgeR. (C) Summary of data
structures for the storage of methylation data used in different parts
of the workflow. Xi: inactive X. Xa: active X. CAST: Castaneus. 5mC:
5-methylcytosine. MDS: multidimensional scaling. DMR: differentially
methylated region. Figure created with BioRender.com.
Loading in data
library(BiocFileCache)
library(dmrseq)
library(dplyr)
library(readr)
library(plyranges)
library(NanoMethViz)
set.seed(1234)We begin by loading the packages and downloading the input data which contains the modBAM files required for this workflow.
if (!dir.exists("input")) {
cache_path <- tools::R_user_dir("LongMod123", which = "cache")
bfc <- BiocFileCache::BiocFileCache(cache_path, ask = FALSE)
url <- "https://zenodo.org/records/12747551/files/input.tar.gz?download=1"
qry <- BiocFileCache::bfcquery(bfc, url, exact = TRUE)
if (nrow(qry) == 0) {
input_path <- BiocFileCache::bfcadd(bfc, "input", fpath = url)
} else {
if (BiocFileCache::bfcneedsupdate(bfc, qry$rid)) {
BiocFileCache::bfcdownload(bfc, qry$rid)
}
input_path <- bfc[[qry$rid]]
}
utils::untar(input_path, exdir = ".")
}As mentioned, modBAM files are the currently preferred output of
Dorado, the primary methylation caller offered by ONT. modBAM files are
BAM files containing reads with additional tags on each read storing
information about methylation calls (the MM tag) alongside their
methylation probabilities (the ML tag). To use this data in
NanoMethViz, we must construct an object of the
ModBamResults class.
The ModBamResults class is a container for storing
methylation data from multiple samples, along with sample annotation and
exon annotation. To create this object, we need to provide paths to the
modBAM files, a sample table, and optionally exon annotation.
# List the modBAM files by searching for files with the .bam extension in the input directory
bam_files <- dir("input", pattern = "*bam$", full.names = TRUE)
bam_files## [1] "input/nsc_1_mat.bam" "input/nsc_1_pat.bam" "input/nsc_2_mat.bam"
## [4] "input/nsc_2_pat.bam" "input/nsc_3_mat.bam" "input/nsc_3_pat.bam"The samples need to be annotated with information for
NanoMethViz to use in aggregating data within experimental
groups. The sample table must contain sample and
group columns with optional additional columns for further
annotation. The rows of this table must match the order of the input
files. The group column is generally the default grouping
column used in the NanoMethViz package. In this example, we
have grouped our data by haplotype.
samples <- read_tsv("input/sample_anno.tsv", show_col_types = FALSE)
samples## # A tibble: 6 × 4
## sample group tissue haplotype
## <chr> <chr> <chr> <chr>
## 1 nsc_1_mat nsc_mat nsc mat
## 2 nsc_1_pat nsc_pat nsc pat
## 3 nsc_2_mat nsc_mat nsc mat
## 4 nsc_2_pat nsc_pat nsc pat
## 5 nsc_3_mat nsc_mat nsc mat
## 6 nsc_3_pat nsc_pat nsc patOptionally we can add exon annotation to the object to generate
tracks for gene annotation when plotting genomic regions/genes. The exon
annotation must be a data.frame with columns
gene_id, chr, strand,
start, end, transcript_id, and
symbol. A number of helper functions are provided to
retrieve exon annotations in the correct format for human (hg19, hg38)
and mouse (mm10, GRCm39). In this example, we will use the
get_exons_mm10() function to retrieve exon annotations for
the mouse genome (mm10) and filter for chromosome 7.
exon_anno <- get_exons_mm10() %>%
dplyr::filter(chr == "chr7")
exon_anno## # A tibble: 60,146 × 7
## gene_id chr strand start end transcript_id symbol
## <chr> <chr> <chr> <int> <int> <int> <chr>
## 1 100009609 chr7 - 84935565 84941088 60883 Vmn2r65
## 2 100009609 chr7 - 84943141 84943264 60883 Vmn2r65
## 3 100009609 chr7 - 84943504 84943722 60883 Vmn2r65
## 4 100009609 chr7 - 84946200 84947000 60883 Vmn2r65
## 5 100009609 chr7 - 84947372 84947651 60883 Vmn2r65
## 6 100009609 chr7 - 84963816 84964115 60883 Vmn2r65
## 7 100009609 chr7 - 84935565 84941088 60884 Vmn2r65
## 8 100009609 chr7 - 84943141 84943264 60884 Vmn2r65
## 9 100009609 chr7 - 84943504 84943722 60884 Vmn2r65
## 10 100009609 chr7 - 84946200 84947000 60884 Vmn2r65
## # ℹ 60,136 more rowsOnce we have created the ModBamResult object, we can use
the plot_gene() function to visualise the methylation
patterns of a gene of interest. In this example, we will plot the
methylation patterns of the Peg3 (paternally-expressed gene 3)
gene on chromosome 7, which is known to be imprinted in mice. The plot
contains a smoothed trendline of the group-aggregated methylation
probabilities across the gene, along with a heatmap showing the
methylation probabilities of individual CpG sites in each read, and a
track showing the isoforms of the gene if exon annotation is provided
(Figure @ref(fig:fig2-gene-peg3)).
mbr <- ModBamResult(
methy = ModBamFiles(
paths = bam_files,
samples = samples$sample
),
samples = samples,
exons = exon_anno,
mod_code = "m"
)
plot_gene(mbr, "Peg3")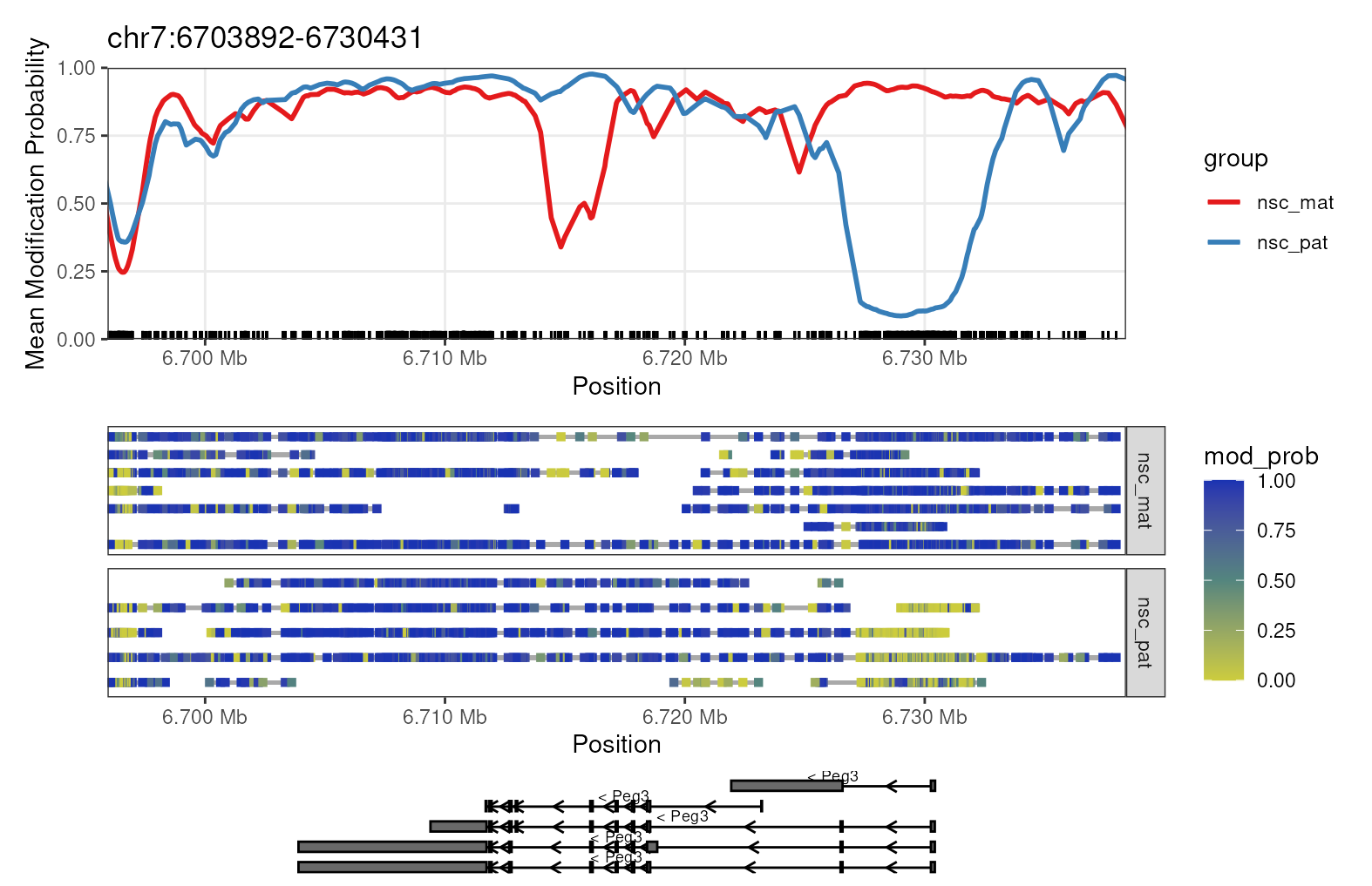
Gene-level plot of Peg3. This plot shows the methylation information along the Peg3 gene on chromosome 7. The top track shows a rolling-smoothed average of methylation probability across the region, aggregated across samples within each experimental group. The middle track shows a heatmap of the methylation probabilities within individual reads, separated by experimental group. The bottom track shows isoform annotations for Peg3, with rectangles representing exons, lines representing introns and arrows representing the direction of transcription.
CpG dinucleotides are often enriched around gene promoters, where methylation is generally correlated with silencing and hypomethylation with activation. In the plot, we see that there is a strong demethylation pattern on the paternal allele near the transcription start site (TSS) of the gene, allowing the gene to be expressed. The maternal allele is hypermethylated near the TSS, silencing the gene. This is a typical methylation pattern for an imprinted gene, and consistent with paternal expression of Peg3.
Genome-wide analysis
While it is possible to use modBAM files to analyse specific regions
of interest, genome-wide analysis requires the data to be in a
tabix-indexed TSV file as it is easier to parse at scale (Figure C). We
can convert the modBAM files to a tabix-indexed TSV file using the
modbam_to_tabix() function. This function will create a
tabix-indexed TSV file containing the methylation data from the modBAM
files. This file can then be used as input to the
NanoMethViz functions that require sorted genomic data. In
this example, we will convert the ModBamResult object to a
tabix-indexed TSV file and save it to the data directory.
We will use a pre-generated file in the interest of time, but you can
run the code below to generate the file.
dir.create("data", showWarnings = FALSE)
if (!file.exists("data/methy.tsv.bgz")) {
modbam_to_tabix(mbr, "data/methy.tsv.bgz")
}Once we have the tabix-indexed TSV file, we can create a
NanoMethResult object using the
NanoMethResult() function. The NanoMethResult
object contains the same information as the ModBamResult
object but with the methylation data stored in a tabix-indexed TSV file.
To create this object we need to provide paths to the tabix-indexed TSV
file, along with the accompanying sample table and exon annotation we
generated before.
nmr <- NanoMethResult(
methy = "data/methy.tsv.bgz",
samples = samples,
exons = exon_anno
)The NanoMethResult object behaves in the same way as
ModBamResult for any plotting functions. For example we can
use plot_gene() to visualise the methylation patterns of
the Peg3 gene in exactly the same way but replacing the
ModBamResult object with the NanoMethResult
object. We should see the exact same plot as before.
plot_gene(nmr, "Peg3")Aggregating feature methylation data
It is often informative to aggregate methylation data over a class of
features to identify broad patterns of methylation across the features.
This can help establish overall differences in methylation patterns
between groups in a class of features such as genes, CpG islands, or
enhancers. The NanoMethViz package provides a
plot_agg_regions() function for visualising methylation
data that has been aggregated across a set of genomic regions of
interest, defined by a table of coordinates.
For example, we may want to investigate the methylation patterns of a
set of genes on chromosome 7. We can use the
exons_to_genes() helper function to convert exon
annotations already stored in the object to gene annotations and filter
for only chromosome 7 genes (Figure @ref(fig:fig3-gene-agg)). For faster
processing, we subset 200 genes and use the
plot_agg_regions() function to visualise the mean
methylation proportions along the sampled genes. By default, the
methylation proportion is calculated using a threshold of 0.5
modification probability.
gene_anno <- exons_to_genes(exons(nmr))
plot_agg_regions(nmr, regions = slice_sample(gene_anno, n = 200), group_col = "group")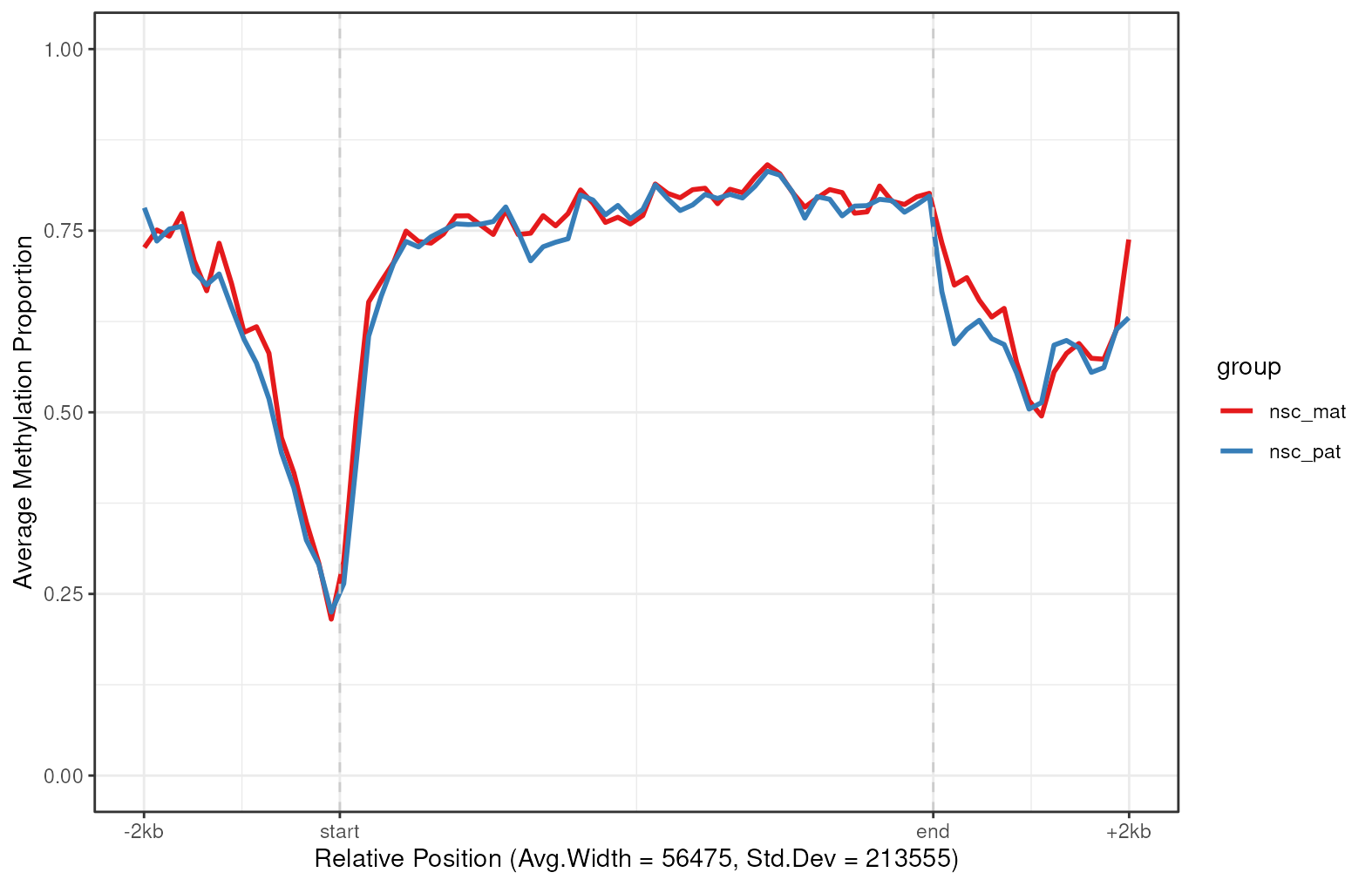
Aggregated gene-level plot. The mean methylation proportion is shown across a sample of 200 genes in chromosome 7, aggregated by experimental group. Methylation proportions are individual CpG sites are calculated along relative genomic positions, with fixed flanking regions, then averaged across all genes in each experimental group to produce a smoothed trendline. The x-axis label gives the average width and standard deviation of widths of the features used.
In this plot we can see that active genes on chromosome 7 tend to be demethylated near the TSS region and hypermethylated throughout the gene body. This is consistent with the proposed function of methylation in gene regulation, where methylation in promoter region of genes is associated with gene silencing, while transcription of active genes is associated with recruitment of methylating mechanisms to the gene body.
Another class of features of interest are CpG islands, which are
regions of the genome that are rich in CpG dinucleotides. CpG islands
are often associated with gene promoters and are predominantly
unmethylated in normal cells (15). We can
use the get_cgi_mm10() function to retrieve CpG island
annotations for the mouse genome (mm10) and filter for chromosome 7. We
can then use the plot_agg_regions() function to visualise
the mean methylation profile across CpG islands on chromosome 7 (Figure
@ref(fig:fig4-cgi-agg)).
cgi_anno <- get_cgi_mm10() %>%
filter(chr == "chr7")
cgi_anno## # A tibble: 1,193 × 14
## bin chr start end gene_id length cpgNum gcNum perCpg perGc obsExp
## <dbl> <chr> <dbl> <dbl> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 609 chr7 3181031 3181267 CpG: 22 236 22 147 18.6 62.3 1
## 2 610 chr7 3303442 3303817 CpG: 28 375 28 249 14.9 66.4 0.69
## 3 610 chr7 3366695 3366942 CpG: 27 247 27 152 21.9 61.5 1.22
## 4 611 chr7 3411983 3412269 CpG: 26 286 26 188 18.2 65.7 0.84
## 5 611 chr7 3414874 3415855 CpG: 87 981 87 518 17.7 52.8 1.28
## 6 611 chr7 3423522 3423834 CpG: 28 312 28 215 17.9 68.9 0.87
## 7 612 chr7 3617298 3617534 CpG: 17 236 17 146 14.4 61.9 0.76
## 8 612 chr7 3629691 3630056 CpG: 32 365 32 195 17.5 53.4 1.24
## 9 612 chr7 3645002 3645963 CpG: 106 961 106 652 22.1 67.8 0.96
## 10 612 chr7 3665238 3665517 CpG: 27 279 27 184 19.4 65.9 0.93
## # ℹ 1,183 more rows
## # ℹ 3 more variables: transcript_id <chr>, strand <chr>, symbol <chr>
plot_agg_regions(nmr, cgi_anno, group_col = "group")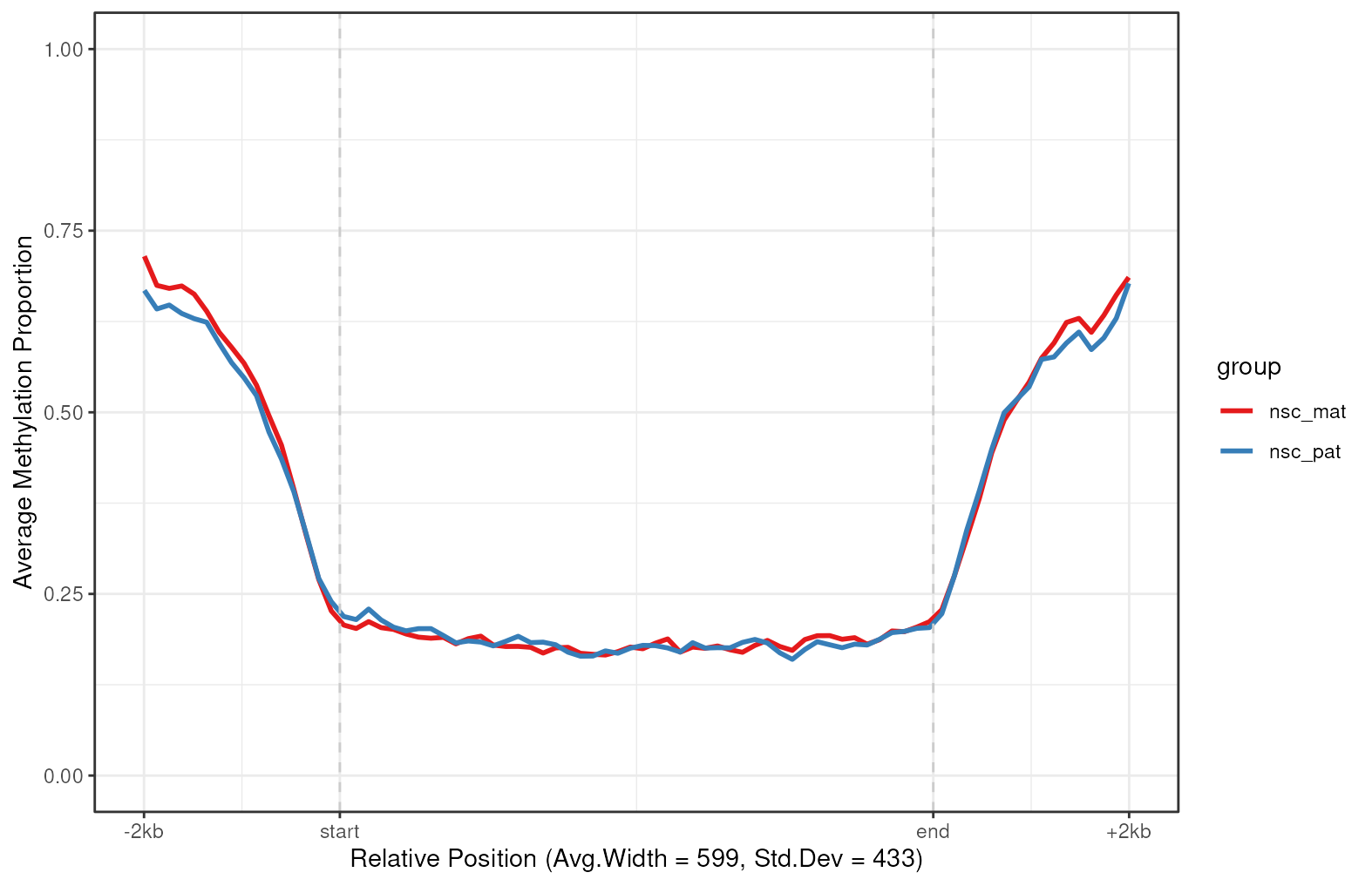
Aggregated CpG island-level plot. The mean methylation proportion is shown across CpG islands on chromosome 7, aggregated by experimental group. Methylation proportions are individual CpG sites are calculated along relative genomic positions, with fixed flanking regions, then averaged across all CpG islands in each experimental group to produce a smoothed trendline.
As expected, we see a clear pattern of demethylation over CpG islands on chromosome 7, relative to neighbouring genomic regions.
Differential methylation
NanoMethViz is primarily designed for visualising
methylation data. In order to perform differential methylation analysis,
we can use other Bioconductor packages that implement sophisticated
statistical procedures, such as dmrseq. The
dmrseq() function requires a BSseq object as
input, so we convert the NanoMethResult object to a
BSseq object using the methy_to_bsseq()
function.
We remove any sites that are zero in all of the samples of either
experimental group; these low coverage regions are generally not
informative for differential methylation analysis. The
BSseq object stores the methylation data as two matrices,
the methylation matrix and the coverage matrix (Figure @ref(fig:fig1)C),
with the option to store additional information such as sample
annotation and genomic regions.
This conversion process usually takes some time, so we will load a
pre-generated BSseq object. The code used to generate the
object is provided below.
if (!file.exists("data/bss.rds")) {
bss <- methy_to_bsseq(nmr)
saveRDS(bss, "data/bss.rds")
} else {
bss <- readRDS("data/bss.rds")
}
pat_cov <- getCoverage(bss)[, which(bss$group == "nsc_pat")]
mat_cov <- getCoverage(bss)[, which(bss$group == "nsc_mat")]
low_cov <- (rowSums(pat_cov == 0) == 3) | (rowSums(mat_cov == 0) == 3)
proportions(table(low_cov))## low_cov
## FALSE TRUE
## 0.2853227 0.7146773
bss <- bss[!low_cov, ]MDS Plot
Using the BSseq object, we are able to create a
multi-dimensional scaling (MDS) plot to visualise the relative
differences in methylation between the samples, a method commonly used
in differential gene expression analysis. The MDS function within
NanoMethViz uses the plotMDS() function from
limma (16). Since
BSseq objects store the methylation data as two matrices,
the methylation matrix and the coverage matrix, we need to convert the
BSseq object such that each CpG site from each sample is
represented by a single value in a matrix. This can be done using the
bsseq_to_log_methy_ratio() function, which converts the
BSseq object to a matrix of the log of the methylation
ratio with a small count added to prevent division by zero.
The plot_mds() function can then be used to create an
MDS plot of the samples using the log methylation ratio matrix as input.
The samples are coloured by group – in this case, maternal and
paternal.
Because the number of CpG sites is large, here we aggregate the
methylation data over CpG islands to both reduce the size of the data
and produce a more stable measure of methylation. This is done by
providing the regions = cgi_anno argument to
bsseq_to_log_methy_ratio(). Without the argument, the
function will use individual CpG sites. After the data is aggregated,
plot_mds() uses only the top 500 most variable features to
create the MDS plot (Figure @ref(fig:fig5-mds)).
lmr <- bsseq_to_log_methy_ratio(
bss,
regions = cgi_anno
)
plot_mds(lmr, groups = bss$group)
MDS plot, The multi-dimensional plot is created based on the top 500 most variable genes in on chromosome 7 in terms of log-methylation-ratio over the gene bodies. The samples are coloured by group, and distances between samples is reflective of the difference in the methylation profile across genes in each sample.
From this MDS, we see that there is clear separation between the maternal and paternal groups based on CpG islands on chromosome 7, suggesting that the most variable CpG islands are differentially methylated between the two groups. Given that this is autosomal data, the separation is likely due to imprinting or strain-specific effects. We therefore expect to find some differentially methylated regions between the maternal and paternal groups.
Differentially methylated regions
Another common analysis is to identify differentially methylated regions (DMRs) between experimental groups. It is often informative to identify DMRs rather than just single differentially-methylated CpG sites, as these may correspond to gene regulatory regions such as promoters, silencers and enhancers, and have biological relevance for differential epigenetic regulation. However, DMRs are not easy to identify, as DMR calling requires the identification of individual significant sites, some determination of whether nearby sites should be aggregated into a region, and a statistical test to determine if the region as a whole is differentially methylated.
The dmrseq package can be used to identify DMRs between
the two groups in our data. The dmrseq() function from the
package requires a BSseq object as input, along with the
name of the covariate to test for differential methylation. We will test
for differential methylation between the maternal and paternal groups.
The dmrseq() function returns a GRanges object
containing the DMRs identified by the function. The GRanges
object contains the genomic coordinates of the DMRs, along with
information about the statistical significance of the DMRs.
There are alternatives to dmrseq for identifying DMRs,
such as bsseq, DSS and edgeR. For
bsseq and DSS, the same BSseq
object can be used as input, while edgeR requires a
DGEList object as input (Figure @ref(fig:fig1)C). The
DGEList object can be created from the BSseq
object using the bsseq_to_edger() function, or from the
NanoMethResult object using the
methy_to_edger() function (Figure @ref(fig:fig1)B).
bsseq, DSS and dmrseq all use
a similar approach to identify DMRs: first, they use a statistical test
to compare the methylation levels between the two groups at each CpG
site, followed by a clustering step to identify regions of the genome
with consistent differences in methylation levels for de novo DMR
discovery. On the other hand, edgeR requires users to
choose the regions to test for differential methylation, which is
reflected by the regions over which they choose to summarise counts, and
therefore cannot be used for de novo DMR discovery. dmrseq
and edgeR produce
-values
for each region, while bsseq and DSS only
provide
-values
for each CpG site along with an aggregated area statistic.
if (!file.exists("data/regions.rds")) {
regions <- dmrseq::dmrseq(bss, testCovariate = "group", minNumRegion = 20)
saveRDS(regions, "data/regions.rds")
} else {
regions <- readRDS("data/regions.rds")
}A number of regions will be produced by dmrseq,
including regions that were aggregated because they contained
significant CpG sites but subsequently failed to reach statistical
significance at a region level. It is difficult to directly interpret
the regions returned by differential methylation analysis, and we
commonly wish to identify these regions are possible control regions for
genes. We do this by associating DMRs with genes using proximity to the
gene transcription start site (TSS), first by constructing an annotation
of a promoter region, then overlapping it with the DMRs. We use a
liberal window of 10kb around the TSS to capture potential promoter
regions, this is done by transforming the gene annotation to a
GRanges object, then using the promoters()
function from IRanges (17) to
define the promoter region as 5kb upstream and 5kb downstream of the
TSS. We then use the join_overlap_intersect() function from
plyranges to find the DMRs that overlap with the gene TSS
regions. This will return a GRanges object containing the
regions where DMRs which overlap with the promoter region, and the
associated gene in that region.
gene_anno_gr <- as(gene_anno, "GRanges")
gene_anno_gr_tss <- IRanges::promoters(gene_anno_gr, upstream = 5000, downstream = 5000)
gene_dmr_overlaps <- plyranges::join_overlap_intersect(regions, gene_anno_gr_tss)
gene_dmr_overlaps## GRanges object with 868 ranges and 9 metadata columns:
## seqnames ranges strand | L area beta
## <Rle> <IRanges> <Rle> | <integer> <numeric> <numeric>
## [1] chr7 6727308-6731838 * | 157 128.8218 -1.84082
## [2] chr7 6727308-6731838 * | 157 128.8218 -1.84082
## [3] chr7 143294268-143296757 * | 112 92.9051 -1.87215
## [4] chr7 60003279-60005228 * | 60 50.9278 -1.82583
## [5] chr7 60003279-60005228 * | 60 50.9278 -1.82583
## ... ... ... ... . ... ... ...
## [864] chr7 126579062-126580889 * | 27 5.00269 0.0232921
## [865] chr7 126580818-126580889 * | 27 5.00269 0.0232921
## [866] chr7 126579942-126580889 * | 27 5.00269 0.0232921
## [867] chr7 19696925-19697636 * | 23 2.65049 -0.0118438
## [868] chr7 19696925-19697636 * | 23 2.65049 -0.0118438
## stat pval qval index gene_id symbol
## <numeric> <numeric> <numeric> <IRanges> <character> <character>
## [1] -40.6010 5.71657e-05 0.0401684 27111-27267 18616 Peg3
## [2] -40.6010 5.71657e-05 0.0401684 27111-27267 57775 Usp29
## [3] -32.5253 1.14331e-04 0.0401684 910828-910939 63830 Kcnq1ot1
## [4] -20.1018 1.14331e-04 0.0401684 335287-335346 52480 Snhg14
## [5] -20.1018 1.14331e-04 0.0401684 335287-335346 84704 Snurf
## ... ... ... ... ... ... ...
## [864] 0.0925951 0.996341 0.997876 764165-764191 102465641 Mir7059
## [865] 0.0925951 0.996341 0.997876 764165-764191 12752 Cln3
## [866] 0.0925951 0.996341 0.997876 764165-764191 171504 Apobr
## [867] -0.0795915 0.996456 0.997876 81437-81459 11812 Apoc1
## [868] -0.0795915 0.996456 0.997876 81437-81459 11816 Apoe
## -------
## seqinfo: 1 sequence from an unspecified genome; no seqlengthsAfter we identify the DMRs that overlap with the gene TSS regions, we can filter for the significant DMRs using the qval column. This is the adjusted -value for the DMRs, calculated using the Benjamini-Hochberg procedure to control false-discovery rates for multiple-testing. We can then select the columns we are interested in, such as the gene symbol, chromosome, start and end positions of the DMRs, the strand of the gene, and the q-value of the DMRs.
dmr_regions <- as_tibble(gene_dmr_overlaps) %>%
dplyr::rename(chr = "seqnames")
signif_regions <- dmr_regions %>%
dplyr::filter(qval < 0.05)
signif_regions %>%
dplyr::select(symbol, chr, start, end, strand, qval)## # A tibble: 8 × 6
## symbol chr start end strand qval
## <chr> <fct> <int> <int> <fct> <dbl>
## 1 Peg3 chr7 6727308 6731838 * 0.0402
## 2 Usp29 chr7 6727308 6731838 * 0.0402
## 3 Kcnq1ot1 chr7 143294268 143296757 * 0.0402
## 4 Snhg14 chr7 60003279 60005228 * 0.0402
## 5 Snurf chr7 60003279 60005228 * 0.0402
## 6 Mkrn3 chr7 62416953 62423032 * 0.0402
## 7 Cdkn1c chr7 143459739 143462038 * 0.0402
## 8 Peg12 chr7 62461469 62464053 * 0.0402We now have a list of DMRs which overlap the TSS of genes on
chromosome 7 and have a q-value < 0.05. The genes on this list are
all known imprinted genes in the mouse, including Peg3, which
we plotted in the first section of this workflow. We can plot the
methylations of these genes and highlight the DMRs by setting the
plot_gene() function’s anno_regions argument
to the regions identified. This will highlight the significant regions
in the gene plot in a shaded band. Here we will plot Peg3,
Kcnq1ot1, and Cdkn1c (Figure
@ref(fig:fig6-dmr-genes)). We can see that for all three of these genes,
as required by our filtering criteria, the differentially methylated
regions fall near the TSS.
options("NanoMethViz.highlight_col" = "green")
(patchwork::wrap_elements(plot_gene(nmr, "Peg3", anno_regions = signif_regions)) /
patchwork::wrap_elements(plot_gene(nmr, "Kcnq1ot1", anno_regions = signif_regions)) /
patchwork::wrap_elements(plot_gene(nmr, "Cdkn1c", anno_regions = signif_regions)))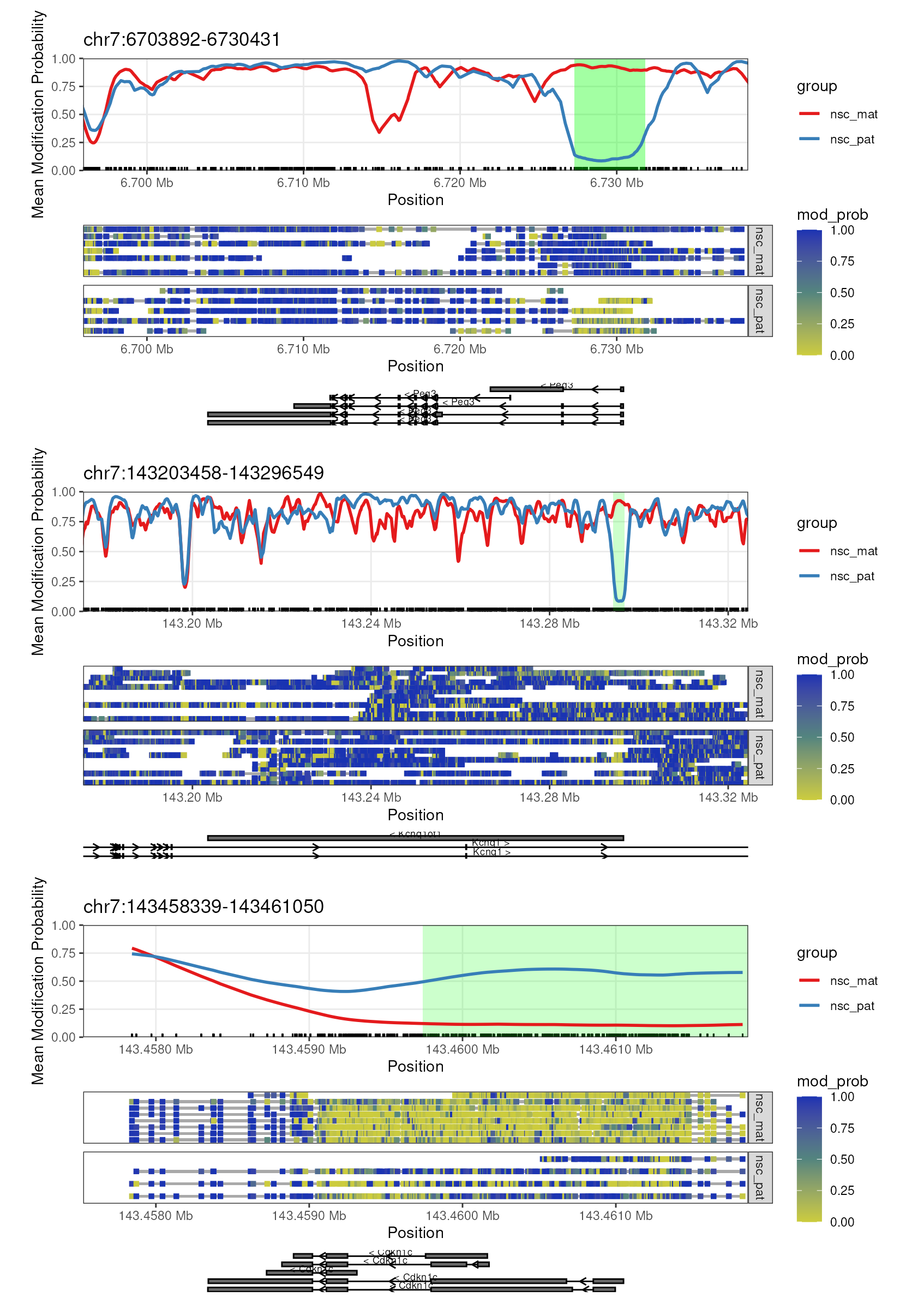
Gene plots with DMR. The plots show the genes Peg3, Kcnq1ot1, and Cdkn1c along with green highlighted region denoting the DMR discovered by dmrseq.
We can see that the DMR for Cdkn1c extends beyond the region
that is plotted when we use plot_gene(). We can plot these
DMRs using plot_region(), with 2000 bases of flanking
sequence added to both sides of the DMR to better visualise the full
scope of the differentially methylated region (Figure
@ref(fig:fig7-cdkn1-expand)).
plot_region(nmr, "chr7", 143459739-2000, 143462038+2000, anno_regions = signif_regions)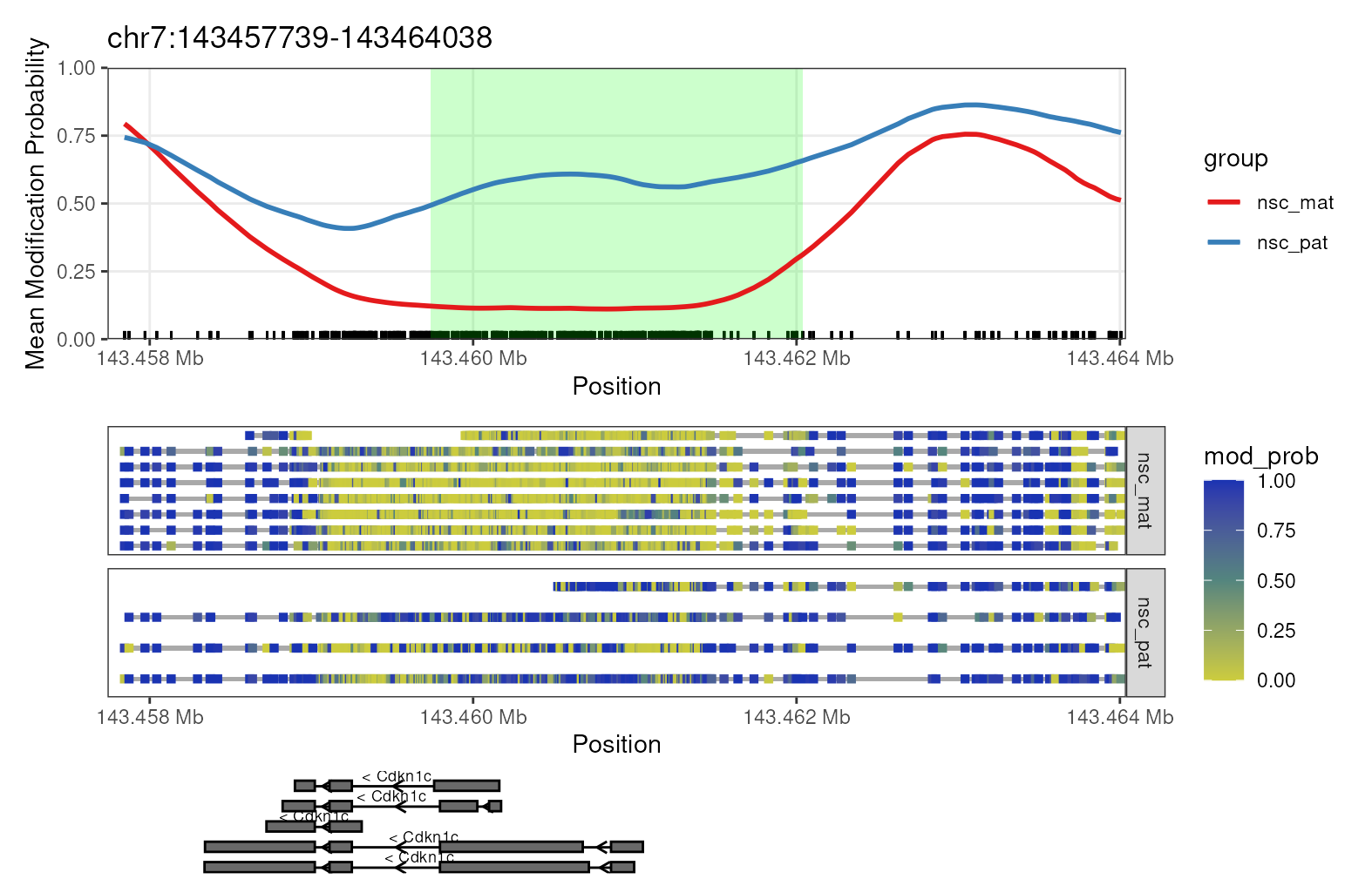
Expanded region around Cdkn1c. This plot shows a more expanded region around the gene Cdkn1c to show the full DMR.
We have now successfully used dmrseq to statistically
identify differentially methylated regions around known imprinted genes.
Additionally, we have used that information to visualise the methylation
data of the genes associated with the DMRs, and identify maternal- and
paternal-specific methylation of the TSS.
Summary
In this workflow, we have demonstrated a complete end-to-end pipeline
for analysing and visualising methylation data generated by ONT
long-read sequencing using NanoMethViz, dmrseq
and other Bioconductor packages. We have shown how to preprocess the
data, load it into NanoMethViz, visualise the methylation
patterns of genes and aggregated features, cluster samples using MDS
plots, and identify DMRs between experimental groups. We have also shown
how to associate DMRs with genes and visualise the methylation patterns
of genes associated with DMRs. While the DMRs identified in this
tutorial were all associated with known imprinted genes, this workflow
could be used in other experimental contexts to identify candidate genes
involved in disease pathogenesis, or discover new gene regulatory
elements. Further analysis could include enrichment analysis of the
differentially-methylated genes, to bring out pathways that may be up-
or downregulated in different processes. Ultimately, this workflow
demonstrates a toolkit for the exploration and analysis of methylation
data from long-read sequencing, which can be applied to a range of
resolution levels and biological contexts. While the workflow was
demonstrated on 5mC ONT nanopore data, it can also be applied to other
long-read DNA methylation data such as that produced by PacBio SMRT
platforms, provided the data is in modBAM format. In principle, it can
be applied to other modification types such as 5hmC or 6mA, as well as
RNA modifications, but this application has not been thoroughly
tested.
Software Avaliability
This workflow makes use of various R packages available from the
Bioconductor project, with version numbers shown below. The
LongMod123 workflow package itself is available at and from
Bioconductor.
## R Under development (unstable) (2025-01-20 r87609)
## Platform: x86_64-pc-linux-gnu
## Running under: Ubuntu 24.04.1 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## time zone: Etc/UTC
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats4 stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] NanoMethViz_3.3.3 ggplot2_3.5.1
## [3] plyranges_1.27.0 readr_2.1.5
## [5] dplyr_1.1.4 dmrseq_1.27.0
## [7] bsseq_1.43.1 SummarizedExperiment_1.37.0
## [9] Biobase_2.67.0 MatrixGenerics_1.19.1
## [11] matrixStats_1.5.0 GenomicRanges_1.59.1
## [13] GenomeInfoDb_1.43.4 IRanges_2.41.2
## [15] S4Vectors_0.45.2 BiocGenerics_0.53.6
## [17] generics_0.1.3 BiocFileCache_2.15.1
## [19] dbplyr_2.5.0
##
## loaded via a namespace (and not attached):
## [1] splines_4.5.0
## [2] BiocIO_1.17.1
## [3] bitops_1.0-9
## [4] filelock_1.0.3
## [5] tibble_3.2.1
## [6] R.oo_1.27.0
## [7] XML_3.99-0.18
## [8] lifecycle_1.0.4
## [9] lattice_0.22-6
## [10] vroom_1.6.5
## [11] magrittr_2.0.3
## [12] limma_3.63.3
## [13] sass_0.4.9
## [14] rmarkdown_2.29
## [15] jquerylib_0.1.4
## [16] yaml_2.3.10
## [17] doRNG_1.8.6.1
## [18] DBI_1.2.3
## [19] RColorBrewer_1.1-3
## [20] abind_1.4-8
## [21] zlibbioc_1.53.0
## [22] purrr_1.0.2
## [23] R.utils_2.12.3
## [24] RCurl_1.98-1.16
## [25] rappdirs_0.3.3
## [26] cpp11_0.5.1
## [27] GenomeInfoDbData_1.2.13
## [28] irlba_2.3.5.1
## [29] pkgdown_2.1.1
## [30] permute_0.9-7
## [31] DelayedMatrixStats_1.29.1
## [32] codetools_0.2-20
## [33] DelayedArray_0.33.4
## [34] tidyselect_1.2.1
## [35] outliers_0.15
## [36] UCSC.utils_1.3.1
## [37] farver_2.1.2
## [38] ScaledMatrix_1.15.0
## [39] GenomicAlignments_1.43.0
## [40] jsonlite_1.8.9
## [41] annotatr_1.33.0
## [42] e1071_1.7-16
## [43] iterators_1.0.14
## [44] systemfonts_1.2.1
## [45] foreach_1.5.2
## [46] dbscan_1.2.2
## [47] tools_4.5.0
## [48] ragg_1.3.3
## [49] Rcpp_1.0.14
## [50] glue_1.8.0
## [51] SparseArray_1.7.4
## [52] mgcv_1.9-1
## [53] xfun_0.50
## [54] HDF5Array_1.35.11
## [55] withr_3.0.2
## [56] BiocManager_1.30.25
## [57] fastmap_1.2.0
## [58] rhdf5filters_1.19.0
## [59] digest_0.6.37
## [60] rsvd_1.0.5
## [61] R6_2.5.1
## [62] textshaping_1.0.0
## [63] colorspace_2.1-1
## [64] Cairo_1.6-2
## [65] gtools_3.9.5
## [66] RSQLite_2.3.9
## [67] R.methodsS3_1.8.2
## [68] h5mread_0.99.4
## [69] utf8_1.2.4
## [70] tidyr_1.3.1
## [71] data.table_1.16.4
## [72] rtracklayer_1.67.0
## [73] class_7.3-23
## [74] httr_1.4.7
## [75] htmlwidgets_1.6.4
## [76] S4Arrays_1.7.1
## [77] org.Mm.eg.db_3.20.0
## [78] regioneR_1.39.0
## [79] pkgconfig_2.0.3
## [80] gtable_0.3.6
## [81] blob_1.2.4
## [82] XVector_0.47.2
## [83] htmltools_0.5.8.1
## [84] scales_1.3.0
## [85] png_0.1-8
## [86] knitr_1.49
## [87] tzdb_0.4.0
## [88] reshape2_1.4.4
## [89] rjson_0.2.23
## [90] nlme_3.1-167
## [91] curl_6.2.0
## [92] bumphunter_1.49.0
## [93] proxy_0.4-27
## [94] cachem_1.1.0
## [95] rhdf5_2.51.2
## [96] stringr_1.5.1
## [97] BiocVersion_3.21.1
## [98] parallel_4.5.0
## [99] vipor_0.4.7
## [100] AnnotationDbi_1.69.0
## [101] ggrastr_1.0.2
## [102] restfulr_0.0.15
## [103] desc_1.4.3
## [104] pillar_1.10.1
## [105] grid_4.5.0
## [106] vctrs_0.6.5
## [107] BiocSingular_1.23.0
## [108] beachmat_2.23.6
## [109] beeswarm_0.4.0
## [110] evaluate_1.0.3
## [111] GenomicFeatures_1.59.1
## [112] cli_3.6.3
## [113] locfit_1.5-9.10
## [114] compiler_4.5.0
## [115] Rsamtools_2.23.1
## [116] rlang_1.1.5
## [117] crayon_1.5.3
## [118] rngtools_1.5.2
## [119] labeling_0.4.3
## [120] plyr_1.8.9
## [121] forcats_1.0.0
## [122] fs_1.6.5
## [123] ggbeeswarm_0.7.2
## [124] TxDb.Mmusculus.UCSC.mm10.knownGene_3.10.0
## [125] stringi_1.8.4
## [126] BiocParallel_1.41.0
## [127] assertthat_0.2.1
## [128] munsell_0.5.1
## [129] Biostrings_2.75.3
## [130] Matrix_1.7-2
## [131] BSgenome_1.75.1
## [132] hms_1.1.3
## [133] patchwork_1.3.0
## [134] sparseMatrixStats_1.19.0
## [135] bit64_4.6.0-1
## [136] Rhdf5lib_1.29.0
## [137] KEGGREST_1.47.0
## [138] statmod_1.5.0
## [139] AnnotationHub_3.15.0
## [140] memoise_2.0.1
## [141] bslib_0.8.0
## [142] bit_4.5.0.1Author contributions
S.S, L.X. and J.L. developed the workflow, performed data analysis, generated figures and wrote the manuscript. T.C., K.B. and M.E.B. generated data analysed in the workflow. P.F.H., M.E.B, Q.G. and M.E.R. supervised the project and wrote the manuscript. All authors read and approved the final manuscript.
Grant information
This work was supported by Australian National Health and Medical Research Council (NHMRC) Investigator Grants (GNT2007996 to Q.G., GNT1194345 to M.E.B. and GNT2017257 to M.E.R), Medical Research Future Fund Researcher Exchange and Development in Industry Fellowship (REDIF249 to M.E.R), Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgements
We thank Kathleen Zeglinski for creating the NanoMethViz
logo and members of the Epigenetics and Development Division at WEHI for
their feedback on this workflow.